Decoding graph neural networks for prediction of alzheimer's disease from neuroimaging and omics datasets
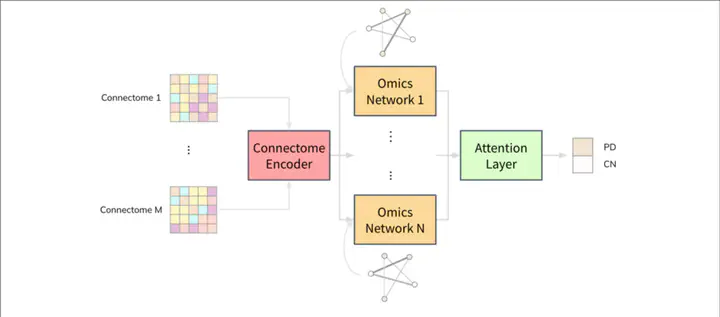 The JOIN-GCLA architecture was origingally developed by researchers from NTU Biomedical Informatics Lab and was used in this project for the classification of AD
The JOIN-GCLA architecture was origingally developed by researchers from NTU Biomedical Informatics Lab and was used in this project for the classification of ADAbstract
AD is referred to as a progressive disease that affects brain cell connections. AD causes brain cells to degenerate and die, eventually destroying memory and other important mental functions. AD is often influenced by genetic factors and the identification of biomarkers can prove useful in the diagnosis of AD. The project aims to predict and carry out decoding to identify structural features of importance. The project aims to deploy the JOIN-GCLA model for classification and uses IG attribution for decoding. sMRI is a non-invasive technique used to analyze the brain to produce illustrations of the brain structure. Multi-omics is a collection of biological data collected from the human body. A new approach was taken to combine multi-Omics Data with sMRI to gain a holistic representation of the human system. The JOIN-GCLA-based model has been trained with both multi-omics and sMRI to detect AD and understand the relevant input features that contribute to AD. This project has implemented the JOIN-GCLA architecture [1] developed by the research group from NTU Biomedical Informatics Lab. The JOIN-GCLA architecture consists of 3 parts: a connectome encoder, omics networks, and an attention layer. The integration of sMRI and multi-omics data to predict AD using the JOIN-GCLA architecture and utilizing Attribution methods such as Integrated Gradients is a novel idea for the identification of biomarkers. The best JOIN-GCLA model from one fold achieved the best accuracy of 96% and an average score of 82.3% with the AnMerge test Data. Decoding the model identified features from the brain that needs to be monitored for predicting AD. Important features identified include the Left-Thalamus, WM-hypointensities, Brain-Stem &, etc.